Comparing networks of different species with seidr¶
Introduction¶
Seidr can compare edges and nodes of two networks that originate from separate species if the user supplies an onthology to translate the node IDs from network A to network B. Consider the these two networks:
net1.sf:
| Source | Target | Type | Weight;Rank |
|---|---|---|---|
| A1 | A2 | Directed | 7.82637e-06;7 |
| A1 | A3 | Directed | 0.131538;5 |
| A2 | A3 | Directed | 0.532767;2 |
| A1 | A4 | Directed | 0.755605;1 |
| A2 | A4 | Directed | 0.218959;4 |
| A1 | A5 | Directed | 0.45865;3 |
| A2 | A5 | Directed | 0.0470446;6 |
net2.sf:
| Source | Target | Type | Weight;Rank |
|---|---|---|---|
| B1 | B2 | Directed | 7.82637e-06;8 |
| B1 | B3 | Directed | 0.131538;6 |
| B2 | B3 | Directed | 0.532767;3 |
| B1 | B4 | Directed | 0.755605;1 |
| B2 | B4 | Directed | 0.218959;5 |
| B1 | B5 | Directed | 0.45865;4 |
| B2 | B5 | Directed | 0.0470446;7 |
| B6 | B7 | Directed | 0.678865;2 |
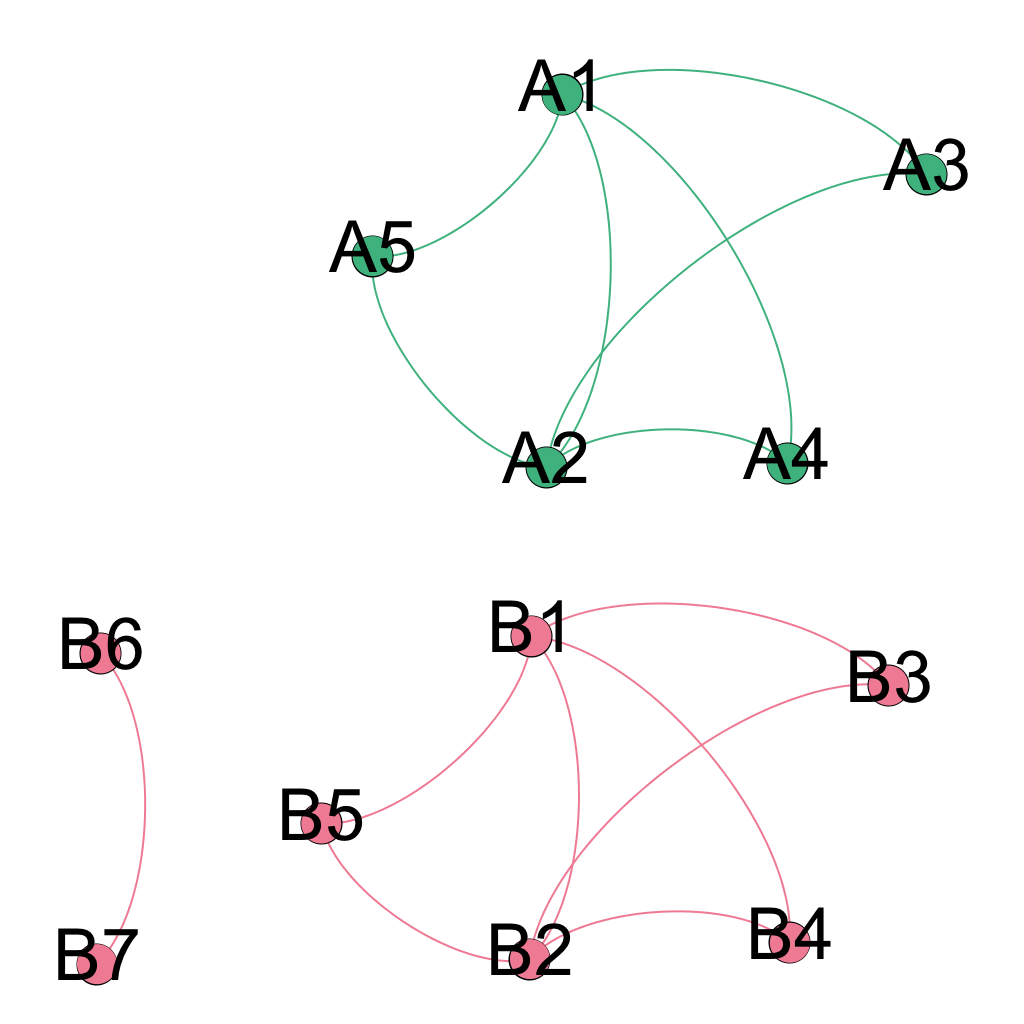
Before we can overlap these two networks, we need to define which nodes are equivalent between them. The file format is a very simple TAB delimited dictionary, each line defining a translation from A to B:
A1 B1
A2 B2
A3 B3
A4 B4
A5 B5
Important info¶
- The compare function currently completely ignores directionality. All output will be undirected.
- There is no support for asymmetric translations. If A1 -> B1, but B1 -> A2 it is left to the user which translation to prioritize.
- Ranks will be merged via \(\sum A_{ij} B_{ij}\) for overlapping edges where \(A_{ij}\) is an edge in network A and \(B_{ij}\) is an edge in network B
- Scores will be computed from all ranks in the dataset via \(\frac{x_i - min(x)}{max(x) - min(x)}\) where \(x\) is a vector of all ranks in the merged network and \(x_i\) is the current rank for edge \(i\)
Running seidr compare¶
As a minimum, the user needs to provide the translation (-t, --translate) and two networks in the binary SeidrFile (see SeidrFiles) format. This will create a new file (by default “compare.sf”) containing the merged network:
seidr compare -t dict.txt net1.sf net2.sf
Output of seidr compare¶
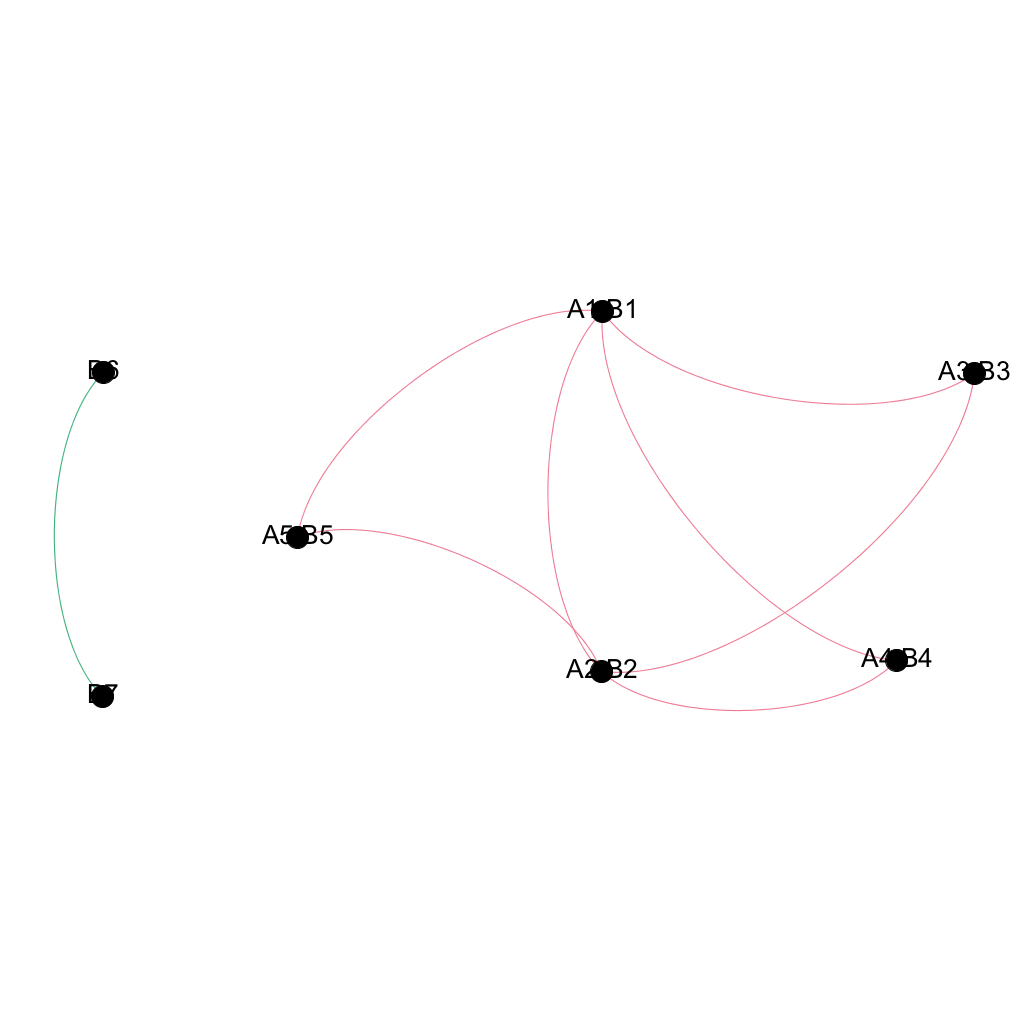
The output of seidr compare in its default mode is a merged network. Nodes with overlaps will be comma separated. If e.g. node A1 ine network A matches node B1 in network B, the joined new node will be “A1,B1”. The fourth column of the merged network contains important metadata for the edges:
- _Flag_: The flag indicates whether the edge was found in both networks (0), only in the first network (1) or only in the second network (2).
- _Rank_A_: This is the original rank of the edge in network A. If it was not present in network A, its rank will be 0.
- _Rank_B_: Analogous to _Rank_A_.
| Source | Target | Type | Weight;Rank | Flag;Rank_A;Rank_B |
|---|---|---|---|---|
| A2,B2 | A1,B1 | Undirected | 0;15 | 0;7;8 |
| A3,B3 | A1,B1 | Undirected | 0.307692;1 | 0;5;6 |
| A3,B3 | A2,B2 | Undirected | 0.769231;5 | 0;2;3 |
| A4,B4 | A1,B1 | Undirected | 1;2 | 0;1;1 |
| A4,B4 | A2,B2 | Undirected | 0.461538;9 | 0;4;5 |
| A5,B5 | A1,B1 | Undirected | 0.615385;7 | 0;3;4 |
| A5,B5 | A2,B2 | Undirected | 0.153846;13 | 0;6;7 |
| B7 | B6 | Undirected | 1;2 | 2;0;2 |
Running seidr compare -n¶
If you are interested in nodes, rather than edges, you can run seidr compare with the -n option. This will create output describing whether a node had at least one edge in network A (1), network B (2) or both (0):
seidr compare -n -t dict.txt net1.sf net2.sf
Output of seidr compare -n¶
The output of seidr compare -n will be 2 columns as TAB delimited text written to stdout indicating whether a node had at least one edge in network A (1), network B (2) or both (0):
A1,B1 0
A2,B2 0
A3,B3 0
A4,B4 0
A5,B5 0
B6 2
B7 2